Rat Malate Dehydrogenase 1 / MDH1 ELISA Kit
- SKU:
- RTFI00951
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- O88989
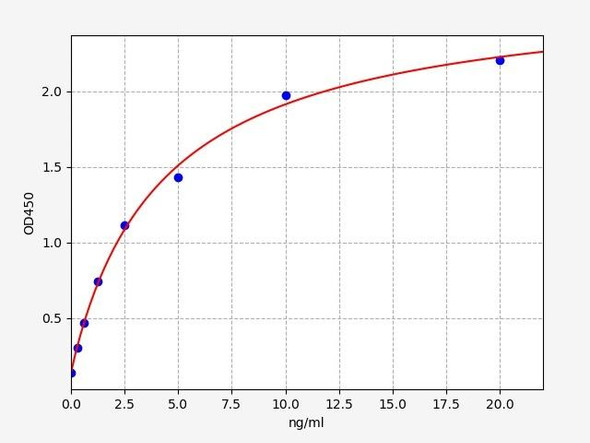
- Sensitivity:
- 46.875pg/ml
- Range:
- 78.125-5000pg/ml
- ELISA Type:
- Sandwich ELISA, Double Antibody
- Synonyms:
- MDH1
- Reactivity:
- Rat
Description
| Product Name: | Rat MDH1 (Malate Dehydrogenase 1) ELISA Kit |
| Product Code: | RTFI00951 |
| Size: | 96 Assays |
| Target: | Rat MDH1 |
| Alias: | MDH1 |
| Reactivity: | Rat |
| Detection Method: | Sandwich ELISA, Double Antibody |
| Sensitivity: | 46.875pg/ml |
| Range: | 78.125-5000pg/ml |
| Storage: | 4°C for 6 months |
| Note: | For Research Use Only |
| Recovery: | Matrices listed below were spiked with certain level of Rat MDH1 and the recovery rates were calculated by comparing the measured value to the expected amount of Rat MDH1 in samples. | ||||||||||||||||
| |||||||||||||||||
| Linearity: | The linearity of the kit was assayed by testing samples spiked with appropriate concentration of Rat MDH1 and their serial dilutions. The results were demonstrated by the percentage of calculated concentration to the expected. | ||||||||||||||||
| |||||||||||||||||
| Intra-Assay: | CV <8% | ||||||||||||||||
| Inter-Assay: | CV <10% |
| Uniprot: | O88989 |
| UniProt Protein Function: | MDH1: Malate dehydrogenase catalyzes the reversible oxidation of malate to oxaloacetate, utilizing the NAD/NADH cofactor system in the citric acid cycle. The protein encoded by this gene is localized to the cytoplasm and may play pivotal roles in the malate-aspartate shuttle that operates in the metabolic coordination between cytosol and mitochondria. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene.[provided by RefSeq, Nov 2010] |
| UniProt Protein Details: | Protein type:EC 1.1.1.37; Carbohydrate Metabolism - glyoxylate and dicarboxylate; EC 1.1.1.96; Carbohydrate Metabolism - pyruvate; Oxidoreductase; Carbohydrate Metabolism - citrate (TCA) cycle Cellular Component: centrosome; cytoplasm; extracellular space; mitochondrion; myelin sheath Molecular Function:L-malate dehydrogenase activity; malate dehydrogenase activity; NAD binding Biological Process: carbohydrate metabolic process; malate metabolic process; NAD metabolic process; NADH metabolic process; oxaloacetate metabolic process; positive regulation of defense response to virus by host; tricarboxylic acid cycle |
| NCBI Summary: | This gene encodes an enzyme that catalyzes the NAD/NADH-dependent, reversible oxidation of malate to oxaloacetate in many metabolic pathways, including the citric acid cycle. Two main isozymes are known to exist in eukaryotic cells: one is found in the mitochondrial matrix and the other in the cytoplasm. This gene encodes the cytosolic isozyme, which plays a key role in the malate-aspartate shuttle that allows malate to pass through the mitochondrial membrane to be transformed into oxaloacetate for further cellular processes. A recent study showed that a C-terminally extended isoform is produced by use of an alternative in-frame translation termination codon via a stop codon readthrough mechanism, and that this isoform is localized in the peroxisomes. [provided by RefSeq, Feb 2016] |
| UniProt Code: | O88989 |
| NCBI GenInfo Identifier: | 15100179 |
| NCBI Gene ID: | 24551 |
| NCBI Accession: | NP_150238.1 |
| UniProt Secondary Accession: | O88989,O88585, Q6PCV2, |
| UniProt Related Accession: | O88989 |
| Molecular Weight: | 36,483 Da |
| NCBI Full Name: | malate dehydrogenase, cytoplasmic isoform Mdh1 |
| NCBI Synonym Full Names: | malate dehydrogenase 1 |
| NCBI Official Symbol: | Mdh1 |
| NCBI Official Synonym Symbols: | MDL1; Mdhl; Mor2 |
| NCBI Protein Information: | malate dehydrogenase, cytoplasmic; malate dehydrogenase, peroxisomal |
| UniProt Protein Name: | Malate dehydrogenase, cytoplasmic |
| UniProt Synonym Protein Names: | Cytosolic malate dehydrogenase |
| Protein Family: | Malate dehydrogenase |
| UniProt Gene Name: | Mdh1 |
| UniProt Entry Name: | MDHC_RAT |
| Step | Procedure |
| 1. | Set standard, test sample and control (zero) wells on the pre-coated plate respectively, and then, record their positions. It is recommended to measure each standard and sample in duplicate. Wash plate 2 times before adding standard, sample and control (zero) wells! |
| 2. | Aliquot 0.1ml standard solutions into the standard wells. |
| 3. | Add 0.1 ml of Sample / Standard dilution buffer into the control (zero) well. |
| 4. | Add 0.1 ml of properly diluted sample ( Human serum, plasma, tissue homogenates and other biological fluids.) into test sample wells. |
| 5. | Seal the plate with a cover and incubate at 37°C for 90 min. |
| 6. | Remove the cover and discard the plate content, clap the plate on the absorbent filter papers or other absorbent material. Do NOT let the wells completely dry at any time. Wash plate X2. |
| 7. | Add 0.1 ml of Biotin- detection antibody working solution into the above wells (standard, test sample & zero wells). Add the solution at the bottom of each well without touching the side wall. |
| 8. | Seal the plate with a cover and incubate at 37°C for 60 min. |
| 9. | Remove the cover, and wash plate 3 times with Wash buffer. Let wash buffer rest in wells for 1 min between each wash. |
| 10. | Add 0.1 ml of SABC working solution into each well, cover the plate and incubate at 37°C for 30 min. |
| 11. | Remove the cover and wash plate 5 times with Wash buffer, and each time let the wash buffer stay in the wells for 1-2 min. |
| 12. | Add 90 µL of TMB substrate into each well, cover the plate and incubate at 37°C in dark within 10-20 min. (Note: This incubation time is for reference use only, the optimal time should be determined by end user.) And the shades of blue can be seen in the first 3-4 wells (with most concentrated standard solutions), the other wells show no obvious color. |
| 13. | Add 50 µL of Stop solution into each well and mix thoroughly. The color changes into yellow immediately. |
| 14. | Read the O.D. absorbance at 450 nm in a microplate reader immediately after adding the stop solution. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum: | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clotovernight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Removeserum and assay promptly or aliquot and store the samples at-80°C. Avoid multiple freeze-thaw cycles. |
| Plasma: | Collect plasma using EDTA or heparin as an anti-coagulant. Centrifuge samples at 4°C for 15 mins at 1000 — g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles.Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid: | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell Culture Supernatant: | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell Lysates: | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20°C. |
| Tissue Homogenates: | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenizein 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or-80°C. |
| Tissue Lysates: | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk: | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |