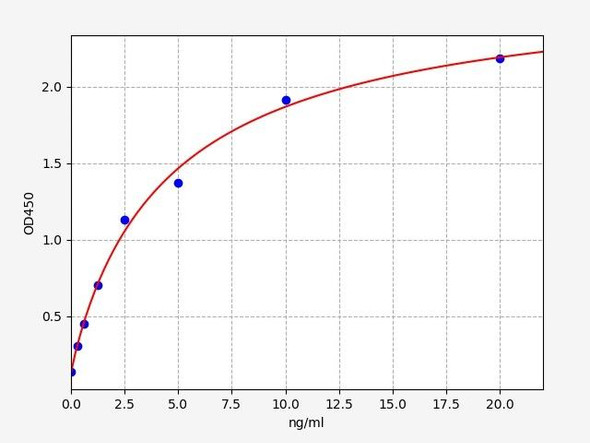
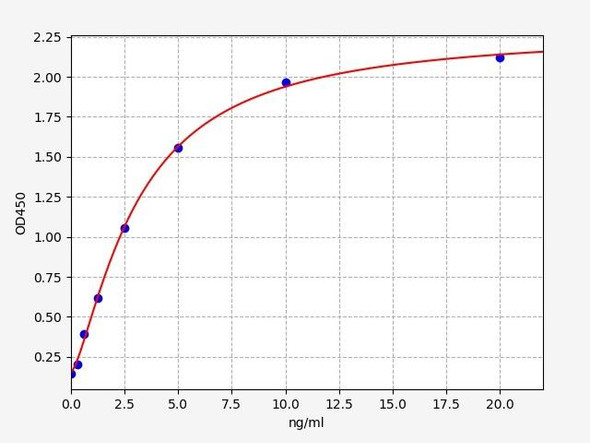
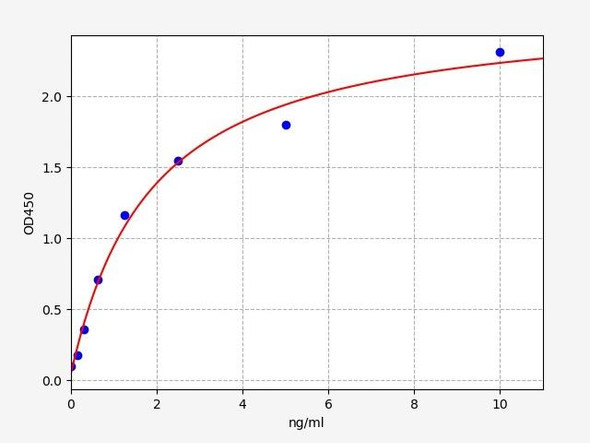
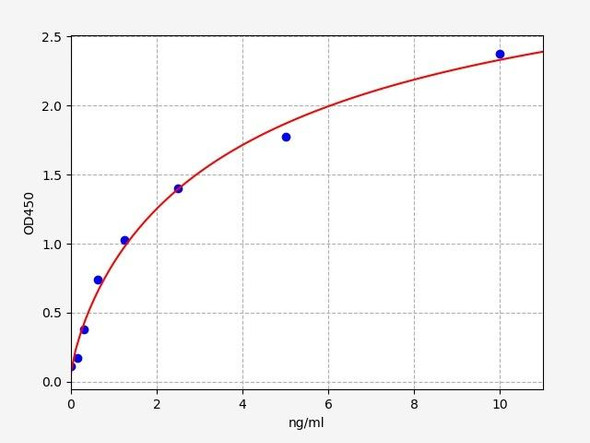
Human Titin (TTN) ELISA Kit
- SKU:
- HUEB1870
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- Q8WZ42
- Range:
- 1.56-100 pmol/mL
- ELISA Type:
- Sandwich
- Synonyms:
- TTN, Titin, Connectin, Rhabdomyosarcoma antigen MU-RMS-40.14, CMD1G, CMH9, CMPD4, EOMFC, HMERF, LGMD2J, MYLK5, TMD
- Reactivity:
- Human
Description
| Product Name: | Human Titin (TTN) ELISA Kit |
| Product Code: | HUEB1870 |
| Alias: | Titin, Connectin, Rhabdomyosarcoma antigen MU-RMS-40.14, TTN, 2.7.11.1 |
| Uniprot: | Q8WZ42 |
| Reactivity: | Human |
| Range: | 1.56-100 pmol/mL |
| Detection Method: | Sandwich |
| Size: | 96 Assay |
| Storage: | Please see kit components below for exact storage details |
| Note: | For research use only |
| UniProt Protein Function: | Function: Key component in the assembly and functioning of vertebrate striated muscles. By providing connections at the level of individual microfilaments, it contributes to the fine balance of forces between the two halves of the sarcomere. The size and extensibility of the cross-links are the main determinants of sarcomere extensibility properties of muscle. In non-muscle cells, seems to play a role in chromosome condensation and chromosome segregation during mitosis. Might link the lamina network to chromatin or nuclear actin, or both during interphase. Ref.28 |
| UniProt Protein Details: | Catalytic activity: ATP + a protein = ADP + a phosphoprotein. Cofactor: Magnesium. Enzyme regulation: Full activation of the protein kinase domain requires both phosphorylation of Tyr-32341, preventing it from blocking the catalytic aspartate residue, and binding of Ca/CALM to the C-terminal regulatory tail of the molecule which results in ATP binding to the kinase. Ref.28 Subunit structure: Interacts with MYOM1, MYOM2, tropomyosin and myosin. Interacts with actin, primarily via the PEVK domains and with MYPN By similarity. Interacts with FHL2, NEB, CRYAB, LMNA/lamin-A and LMNB/lamin-B. Interacts with TCAP/telethonin and/or ANK1 isoform Mu17/ank15, via the first two N-terminal immunoglobulin domains. Interacts with TRIM63 and TRIM55, through several domains including immunoglobulin domains 141 and 142. Interacts with ANKRD1, ANKRD2 and ANKRD23, via the region between immunoglobulin domains 77 and 78 and interacts with CAPN3, via immunoglobulin domain 79. Interacts with NBR1 through the protein kinase domain. Interacts with CALM/calmodulin. Isoform 6 interacts with OBSCN isoform 3. Interacts with CMYA5. Ref.3 Ref.16 Ref.17 Ref.18 Ref.19 Ref.20 Ref.21 Ref.22 Ref.23 Ref.26 Ref.28 Ref.38 Subcellular location: Cytoplasm Probable. Nucleus Ref.23. Tissue specificity: Isoforms 3, 7 and 8 are expressed in cardiac muscle. Isoform 4 is expressed in vertebrate skeletal muscle. Isoform 6 is expressed in skeletal muscle (at protein level). Ref.3 Ref.7 Domain: ZIS1 and ZIS5 regions contain multiple SPXR consensus sites for ERK- and CDK-like protein kinases as well as multiple SP motifs. ZIS1 could adopt a closed conformation which would block the TCAP-binding site.The PEVK region may serve as an entropic spring of a chain of structural folds and may also be an interaction site to other myofilament proteins to form interfilament connectivity in the sarcomere. Post-translational modification: Autophosphorylated By similarity. Ref.14 Ref.28 Involvement in Disease: Hereditary myopathy with early respiratory failure (HMERF) [MIM:603689]: Autosomal dominant, adult-onset myopathy with early respiratory muscle involvement.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.38Cardiomyopathy, familial hypertrophic 9 (CMH9) [MIM:613765]: A hereditary heart disorder characterized by ventricular hypertrophy, which is usually asymmetric and often involves the interventricular septum. The symptoms include dyspnea, syncope, collapse, palpitations, and chest pain. They can be readily provoked by exercise. The disorder has inter- and intrafamilial variability ranging from benign to malignant forms with high risk of cardiac failure and sudden cardiac death.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.32Cardiomyopathy, dilated 1G (CMD1G) [MIM:604145]: A disorder characterized by ventricular dilation and impaired systolic function, resulting in congestive heart failure and arrhythmia. Patients are at risk of premature death.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.34 Ref.35 Ref.37Tardive tibial muscular dystrophy (TMD) [MIM:600334]: Autosomal dominant, late-onset distal myopathy. Muscle weakness and atrophy are usually confined to the anterior compartment of the lower leg, in particular the tibialis anterior muscle. Clinical symptoms usually occur at age 35-45 years or much later.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.33 Ref.36Limb-girdle muscular dystrophy 2J (LGMD2J) [MIM:608807]: An autosomal recessive degenerative myopathy characterized by progressive weakness of the pelvic and shoulder girdle muscles. Severe disability is observed within 20 years of onset.Note: The disease is caused by mutations affecting the gene represented in this entry.Early-onset myopathy with fatal cardiomyopathy (EOMFC) [MIM:611705]: Early-onset myopathies are inherited muscle disorders that manifest typically from birth or infancy with hypotonia, muscle weakness, and delayed motor development. EOMFC is a titinopathy that, in contrast with the previously described examples, involves both heart and skeletal muscle, has a congenital onset, and is purely recessive. This phenotype is due to homozygous out-of-frame TTN deletions, which lead to a total absence of titin's C-terminal end from striated muscles and to secondary CAPN3 depletion.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.39 Miscellaneous: In some isoforms, after the PEVK repeat region there is a long PEVK duplicated region. On account of this region, it has been very difficult to sequence the whole protein. The length of this region (ranging from 183 to 2174 residues), may be a key elastic element of titin. Sequence similarities: Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family.Contains 132 fibronectin type-III domains.Contains 152 Ig-like (immunoglobulin-like) domains.Contains 19 Kelch repeats.Contains 1 protein kinase domain.Contains 17 RCC1 repeats.Contains 14 TPR repeats.Contains 15 WD repeats. Sequence caution: The sequence AAH58824.1 differs from that shown. Reason: Contaminating sequence. Potential poly-A sequence starting in position 553.The sequence AAH70170.1 differs from that shown. Reason: Contaminating sequence. Potential poly-A sequence starting in position 627.The sequence CAA62188.1 differs from that shown. Reason: Frameshift at positions 17036 and 17043. The sequence CAD12455.1 differs from that shown. Reason: Frameshift at positions 17036 and 17043. |
| NCBI Summary: | This gene encodes a large abundant protein of striated muscle. The product of this gene is divided into two regions, a N-terminal I-band and a C-terminal A-band. The I-band, which is the elastic part of the molecule, contains two regions of tandem immunoglobulin domains on either side of a PEVK region that is rich in proline, glutamate, valine and lysine. The A-band, which is thought to act as a protein-ruler, contains a mixture of immunoglobulin and fibronectin repeats, and possesses kinase activity. An N-terminal Z-disc region and a C-terminal M-line region bind to the Z-line and M-line of the sarcomere, respectively, so that a single titin molecule spans half the length of a sarcomere. Titin also contains binding sites for muscle associated proteins so it serves as an adhesion template for the assembly of contractile machinery in muscle cells. It has also been identified as a structural protein for chromosomes. Alternative splicing of this gene results in multiple transcript variants. Considerable variability exists in the I-band, the M-line and the Z-disc regions of titin. Variability in the I-band region contributes to the differences in elasticity of different titin isoforms and, therefore, to the differences in elasticity of different muscle types. Mutations in this gene are associated with familial hypertrophic cardiomyopathy 9, and autoantibodies to titin are produced in patients with the autoimmune disease scleroderma. [provided by RefSeq, Feb 2012] |
| UniProt Code: | Q8WZ42 |
| NCBI GenInfo Identifier: | 384872704 |
| NCBI Gene ID: | 7273 |
| NCBI Accession: | Q8WZ42.4 |
| UniProt Secondary Accession: | |
| UniProt Related Accession: | Q8WZ42 |
| Molecular Weight: | |
| NCBI Full Name: | Titin |
| NCBI Synonym Full Names: | titin |
| NCBI Official Symbol: | TTNÂ Â |
| NCBI Official Synonym Symbols: | TMD; CMH9; CMD1G; CMPD4; EOMFC; HMERF; MYLK5; SALMY; LGMD2J; LGMDR10Â Â |
| NCBI Protein Information: | titin |
| UniProt Protein Name: | Titin |
| UniProt Synonym Protein Names: | Connectin; Rhabdomyosarcoma antigen MU-RMS-40.14 |
| Protein Family: | Titin |
| UniProt Gene Name: | TTNÂ Â |
| UniProt Entry Name: | TITIN_HUMAN |
| Component | Quantity (96 Assays) | Storage |
| ELISA Microplate (Dismountable) | 8×12 strips | -20°C |
| Lyophilized Standard | 2 | -20°C |
| Sample Diluent | 20ml | -20°C |
| Assay Diluent A | 10mL | -20°C |
| Assay Diluent B | 10mL | -20°C |
| Detection Reagent A | 120µL | -20°C |
| Detection Reagent B | 120µL | -20°C |
| Wash Buffer | 30mL | 4°C |
| Substrate | 10mL | 4°C |
| Stop Solution | 10mL | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipette tips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
*Note: The below protocol is a sample protocol. Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Allow all reagents to reach room temperature (Please do not dissolve the reagents at 37°C directly). All the reagents should be mixed thoroughly by gently swirling before pipetting. Avoid foaming. Keep appropriate numbers of strips for 1 experiment and remove extra strips from microtiter plate. Removed strips should be resealed and stored at -20°C until the kits expiry date. Prepare all reagents, working standards and samples as directed in the previous sections. Please predict the concentration before assaying. If values for these are not within the range of the standard curve, users must determine the optimal sample dilutions for their experiments. We recommend running all samples in duplicate.
| Step | |
| 1. | Add Sample: Add 100µL of Standard, Blank, or Sample per well. The blank well is added with Sample diluent. Solutions are added to the bottom of micro ELISA plate well, avoid inside wall touching and foaming as possible. Mix it gently. Cover the plate with sealer we provided. Incubate for 120 minutes at 37°C. |
| 2. | Remove the liquid from each well, don't wash. Add 100µL of Detection Reagent A working solution to each well. Cover with the Plate sealer. Gently tap the plate to ensure thorough mixing. Incubate for 1 hour at 37°C. Note: if Detection Reagent A appears cloudy warm to room temperature until solution is uniform. |
| 3. | Aspirate each well and wash, repeating the process three times. Wash by filling each well with Wash Buffer (approximately 400µL) (a squirt bottle, multi-channel pipette,manifold dispenser or automated washer are needed). Complete removal of liquid at each step is essential. After the last wash, completely remove remaining Wash Buffer by aspirating or decanting. Invert the plate and pat it against thick clean absorbent paper. |
| 4. | Add 100µL of Detection Reagent B working solution to each well. Cover with the Plate sealer. Incubate for 60 minutes at 37°C. |
| 5. | Repeat the wash process for five times as conducted in step 3. |
| 6. | Add 90µL of Substrate Solution to each well. Cover with a new Plate sealer and incubate for 10-20 minutes at 37°C. Protect the plate from light. The reaction time can be shortened or extended according to the actual color change, but this should not exceed more than 30 minutes. When apparent gradient appears in standard wells, user should terminatethe reaction. |
| 7. | Add 50µL of Stop Solution to each well. If color change does not appear uniform, gently tap the plate to ensure thorough mixing. |
| 8. | Determine the optical density (OD value) of each well at once, using a micro-plate reader set to 450 nm. User should open the micro-plate reader in advance, preheat the instrument, and set the testing parameters. |
| 9. | After experiment, store all reagents according to the specified storage temperature respectively until their expiry. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |