Human PARK2(E3 ubiquitin-protein ligase parkin) ELISA Kit
- SKU:
- HUFI03035
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- O60260
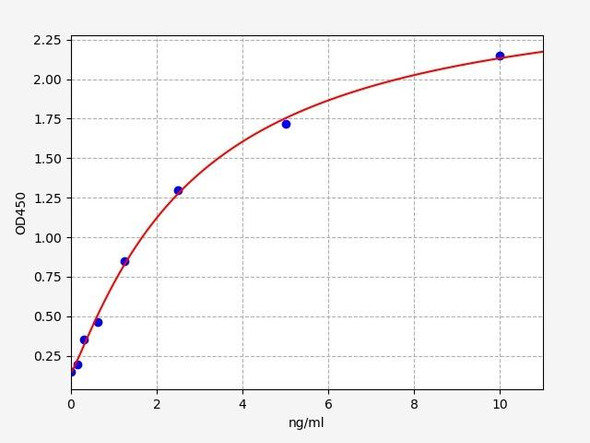
- Sensitivity:
- 0.094ng/ml
- Range:
- 0.156-10ng/ml
- ELISA Type:
- Sandwich ELISA, Double Antibody
- Synonyms:
- E3 ubiquitin-protein ligase parkin
- Reactivity:
- Human
Description
| Product Name: | Human PARK2(E3 ubiquitin-protein ligase parkin) ELISA Kit |
| Product Code: | HUFI03035 |
| Size: | 96 Assays |
| Alias: | E3 ubiquitin-protein ligase parkin |
| Detection method: | Sandwich ELISA, Double Antibody |
| Application: | This immunoassay kit allows for the in vitro quantitative determination of Human PARK2 concentrations in serum plasma and other biological fluids. |
| Sensitivity: | 0.094ng/ml |
| Range: | 0.156-10ng/ml |
| Storage: | 4°C for 6 months |
| Note: | For Research Use Only |
| Recovery: | Matrices listed below were spiked with certain level of Human PARK2 and the recovery rates were calculated by comparing the measured value to the expected amount of Human PARK2 in samples. | ||||||||||||||||
| |||||||||||||||||
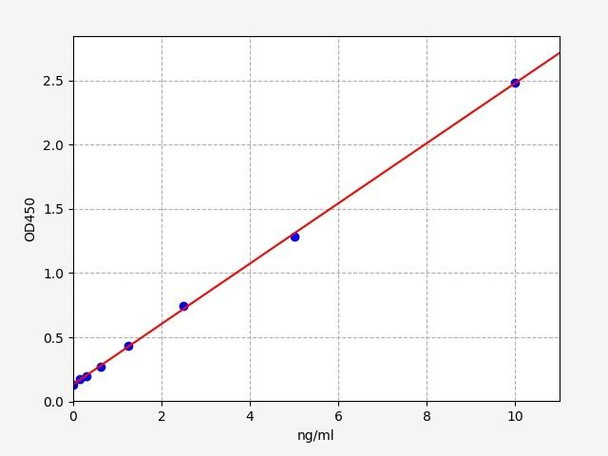
| Linearity: | The linearity of the kit was assayed by testing samples spiked with appropriate concentration of Human PARK2 and their serial dilutions. The results were demonstrated by the percentage of calculated concentration to the expected. | ||||||||||||||||
| |||||||||||||||||
| CV(%): | Intra-Assay: CV<8% Inter-Assay: CV<10% |
| Component | Quantity | Storage |
| ELISA Microplate (Dismountable) | 8×12 strips | 4°C for 6 months |
| Lyophilized Standard | 2 | 4°C/-20°C |
| Sample/Standard Dilution Buffer | 20ml | 4°C |
| Biotin-labeled Antibody(Concentrated) | 120ul | 4°C (Protect from light) |
| Antibody Dilution Buffer | 10ml | 4°C |
| HRP-Streptavidin Conjugate(SABC) | 120ul | 4°C (Protect from light) |
| SABC Dilution Buffer | 10ml | 4°C |
| TMB Substrate | 10ml | 4°C (Protect from light) |
| Stop Solution | 10ml | 4°C |
| Wash Buffer(25X) | 30ml | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipette tips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
| Uniprot | O60260 |
| UniProt Protein Function: | PARK2: a component of a multiprotein E3 ubiquitin ligase complex, catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins, such as BCL2, SYT11, CCNE1, GPR37, STUB1, a 22 kDa O-linked glycosylated isoform of SNCAIP, SEPT5, ZNF746 and AIMP2. Mediates monoubiquitination as well as 'Lys-48'-linked and 'Lys-63'-linked polyubiquitination of substrates depending on the context. Participates in the removal and/or detoxification of abnormally folded or damaged protein by mediating 'Lys-63'-linked polyubiquitination of misfolded proteins such as PARK7: 'Lys-63'- linked polyubiquitinated misfolded proteins are then recognized by HDAC6, leading to their recruitment to aggresomes, followed by degradation. Mediates 'Lys-63'-linked polyubiquitination of SNCAIP, possibly playing a role in Lewy-body formation. Mediates monoubiquitination of BCL2, thereby acting as a positive regulator of autophagy. Promotes the autophagic degradation of dysfunctional depolarized mitochondria. Mediates 'Lys-48'-linked polyubiquitination of ZNF746, followed by degradation of ZNF746 by the proteasome; possibly playing a role in role in regulation of neuron death. Limits the production of reactive oxygen species (ROS). Loss of this ubiquitin ligase activity appears to be the mechanism underlying pathogenesis of PARK2. May protect neurons against alpha synuclein toxicity, proteasomal dysfunction, GPR37 accumulation, and kainate-induced excitotoxicity. May play a role in controlling neurotransmitter trafficking at the presynaptic terminal and in calcium-dependent exocytosis. Regulates cyclin-E during neuronal apoptosis. May represent a tumor suppressor gene. Forms an E3 ubiquitin ligase complex with UBE2L3 or UBE2L6. Mediates 'Lys-63'-linked polyubiquitination by associating with UBE2V1. Part of a SCF-like complex, consisting of PARK2, CUL1 and FBXW7. Part of a complex, including STUB1, HSP70 and GPR37. The amount of STUB1 in the complex increases during ER stress. STUB1 promotes the dissociation of HSP70 from PARK2 and GPR37, thus facilitating PARK2-mediated GPR37 ubiquitination. HSP70 transiently associates with unfolded GPR37 and inhibits the E3 activity of PARK2, whereas, STUB1 enhances the E3 activity of PARK2 through promotion of dissociation of HSP70 from PARK2-GPR37 complexes. Interacts with PSMD4 and PACRG. Interacts with LRRK2. Interacts with RANBP2. Interacts with SUMO1 but not SUMO2, which promotes nuclear localization and autoubiquitination. Interacts (via first RING- type domain) with AIMP2 (via N-terminus). Interacts with PSMA7 and RNF41. Interacts with PINK1. Highly expressed in the brain including the substantia nigra. Expressed in heart, testis and skeletal muscle. Expression is down-regulated or absent in tumor biopsies, and absent in the brain of PARK2 patients. Overexpression protects dopamine neurons from kainate-mediated apoptosis. Found in serum. Belongs to the RBR family. Parkin subfamily. 6 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:Ubiquitin ligase; Ligase; Ubiquitin conjugating system; EC 6.3.2.19; EC 6.3.2.- Chromosomal Location of Human Ortholog: 6q25.2-q27 Cellular Component: Golgi apparatus; neuron projection; mitochondrion; perinuclear region of cytoplasm; endoplasmic reticulum; cytoplasm; SCF ubiquitin ligase complex; nucleus; cytosol; ubiquitin ligase complex Molecular Function:tubulin binding; identical protein binding; ubiquitin binding; zinc ion binding; histone deacetylase binding; ubiquitin-protein ligase activity; Hsp70 protein binding; actin binding; protein kinase binding; PDZ domain binding; protein binding; G-protein-coupled receptor binding; ubiquitin conjugating enzyme binding; chaperone binding; ubiquitin protein ligase binding; heat shock protein binding; kinase binding; SH3 domain binding; ligase activity Biological Process: protein monoubiquitination; proteasomal ubiquitin-dependent protein catabolic process; negative regulation of JNK cascade; negative regulation of actin filament bundle formation; startle response; central nervous system development; protein polyubiquitination; adult locomotory behavior; regulation of protein ubiquitination; protein ubiquitination during ubiquitin-dependent protein catabolic process; regulation of neurotransmitter secretion; protein ubiquitination; mitochondrion localization; norepinephrine metabolic process; dopamine metabolic process; regulation of dopamine secretion; negative regulation of insulin secretion; negative regulation of glucokinase activity; zinc ion homeostasis; negative regulation of protein amino acid phosphorylation; regulation of lipid transport; dopamine uptake; negative regulation of neuron apoptosis; positive regulation of DNA binding; mitochondrion organization and biogenesis; mitochondrial fission; synaptic transmission, glutamatergic; protein autoubiquitination; positive regulation of I-kappaB kinase/NF-kappaB cascade; transcription, DNA-dependent; protein stabilization; learning; regulation of protein transport; cellular protein catabolic process; positive regulation of proteasomal ubiquitin-dependent protein catabolic process; mitochondrion degradation; positive regulation of transcription from RNA polymerase II promoter; regulation of autophagy; response to oxidative stress Disease: Parkinson Disease 2, Autosomal Recessive Juvenile; Leprosy, Susceptibility To, 2; Lung Cancer; Ovarian Cancer |
| NCBI Summary: | The precise function of this gene is unknown; however, the encoded protein is a component of a multiprotein E3 ubiquitin ligase complex that mediates the targeting of substrate proteins for proteasomal degradation. Mutations in this gene are known to cause Parkinson disease and autosomal recessive juvenile Parkinson disease. Alternative splicing of this gene produces multiple transcript variants encoding distinct isoforms. Additional splice variants of this gene have been described but currently lack transcript support. [provided by RefSeq, Jul 2008] |
| UniProt Code: | O60260 |
| NCBI GenInfo Identifier: | 116242725 |
| NCBI Gene ID: | 5071 |
| NCBI Accession: | O60260.2 |
| UniProt Secondary Accession: | O60260,Q5TFV8, Q5VVX4, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44 Q8WW07, A3FG77, A8K975, D3JZW7, D3K2X0, |
| UniProt Related Accession: | O60260 |
| Molecular Weight: | 465 |
| NCBI Full Name: | E3 ubiquitin-protein ligase parkin |
| NCBI Synonym Full Names: | parkin RBR E3 ubiquitin protein ligase |
| NCBI Official Symbol: | PARK2Â Â |
| NCBI Official Synonym Symbols: | PDJ; PRKN; AR-JP; LPRS2Â Â |
| NCBI Protein Information: | E3 ubiquitin-protein ligase parkin; parkinson juvenile disease protein 2; parkinson protein 2, E3 ubiquitin protein ligase (parkin); Parkinson disease (autosomal recessive, juvenile) 2, parkin |
| UniProt Protein Name: | E3 ubiquitin-protein ligase parkin |
| UniProt Synonym Protein Names: | Parkinson juvenile disease protein 2; Parkinson disease protein 2 |
| Protein Family: | E3 ubiquitin-protein ligase |
| UniProt Gene Name: | PARK2Â Â |
| UniProt Entry Name: | PRKN2_HUMAN |
*Note: Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Before adding to wells, equilibrate the SABC working solution and TMB substrate for at least 30 min at 37°C. When diluting samples and reagents, they must be mixed completely and evenly. It is recommended to plot a standard curve for each test.
| Step | Protocol |
| 1. | Set standard, test sample and control (zero) wells on the pre-coated plate respectively, and then, record their positions. It is recommended to measure each standard and sample in duplicate. Wash plate 2 times before adding standard, sample and control (zero) wells! |
| 2. | Aliquot 0.1ml standard solutions into the standard wells. |
| 3. | Add 0.1 ml of Sample / Standard dilution buffer into the control (zero) well. |
| 4. | Add 0.1 ml of properly diluted sample ( Human serum, plasma, tissue homogenates and other biological fluids.) into test sample wells. |
| 5. | Seal the plate with a cover and incubate at 37 °C for 90 min. |
| 6. | Remove the cover and discard the plate content, clap the plate on the absorbent filter papers or other absorbent material. Do NOT let the wells completely dry at any time. Wash plate X2. |
| 7. | Add 0.1 ml of Biotin- detection antibody working solution into the above wells (standard, test sample & zero wells). Add the solution at the bottom of each well without touching the side wall. |
| 8. | Seal the plate with a cover and incubate at 37°C for 60 min. |
| 9. | Remove the cover, and wash plate 3 times with Wash buffer. Let wash buffer rest in wells for 1 min between each wash. |
| 10. | Add 0.1 ml of SABC working solution into each well, cover the plate and incubate at 37°C for 30 min. |
| 11. | Remove the cover and wash plate 5 times with Wash buffer, and each time let the wash buffer stay in the wells for 1-2 min. |
| 12. | Add 90 µl of TMB substrate into each well, cover the plate and incubate at 37°C in dark within 10-20 min. (Note: This incubation time is for reference use only, the optimal time should be determined by end user.) And the shades of blue can be seen in the first 3-4 wells (with most concentrated standard solutions), the other wells show no obvious color. |
| 13. | Add 50 µl of Stop solution into each well and mix thoroughly. The color changes into yellow immediately. |
| 14. | Read the O.D. absorbance at 450 nm in a microplate reader immediately after adding the stop solution. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |