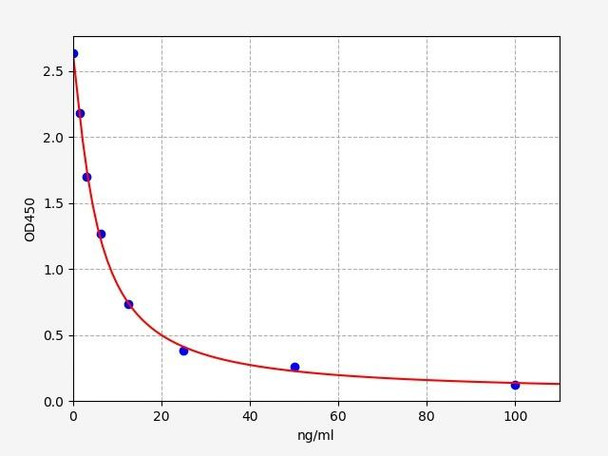
Human HLA-DRB5 / HLA class II histocompatibility , DR beta 5 chain ELISA Kit
- SKU:
- HUFI01440
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- Q30154
- Sensitivity:
- 0.938ng/ml
- Range:
- 1.563-100ng/ml
- ELISA Type:
- Competitive ELISA, Coated with Antigen
- Synonyms:
- HLA-DRB5, HLA class II histocompatibility antigen, DR beta 5 chain, MHC class II antigen DRB5, Dw2, DR beta-5, DR2-beta-2, HLA-DRB
- Reactivity:
- Human
Description
| Product Name: | Human HLA-DRB5 / HLA class II histocompatibility , DR beta 5 chain ELISA Kit |
| Product Code: | HUFI01440 |
| Size: | 96 Assays |
| Alias: | HLA-DRB5, HLA class II histocompatibility antigen, DR beta 5 chain, MHC class II antigen DRB5, Dw2, DR beta-5, DR2-beta-2, HLA-DRB |
| Detection method: | Competitive ELISA, Coated with Antibody |
| Application: | This immunoassay kit allows for the in vitro quantitative determination of Human HLA-DRB5 concentrations in serum plasma and other biological fluids. |
| Sensitivity: | 0.938ng/ml |
| Range: | 1.56-100ng/ml |
| Storage: | 4°C for 6 months |
| Note: | For Research Use Only |
| Recovery: | Matrices listed below were spiked with certain level of Human HLA-DRB5 and the recovery rates were calculated by comparing the measured value to the expected amount of Human HLA-DRB5 in samples. | ||||||||||||||||
| |||||||||||||||||
| Linearity: | The linearity of the kit was assayed by testing samples spiked with appropriate concentration of Human HLA-DRB5 and their serial dilutions. The results were demonstrated by the percentage of calculated concentration to the expected. | ||||||||||||||||
| |||||||||||||||||
| CV(%): | Intra-Assay: CV<8% Inter-Assay: CV<10% |
| Component | Quantity | Storage |
| ELISA Microplate(Dismountable) | 8×12 strips | 4°C for 6 months |
| Lyophilized Standard | 2 | 4°C/-20°C |
| Sample/Standard Dilution Buffer | 20ml | 4°C |
| Biotin-labeled Antibody(Concentrated) | 60ul | 4°C (Protect from light) |
| Antibody Dilution Buffer | 10ml | 4°C |
| HRP-Streptavidin Conjugate(SABC) | 120ul | 4°C (Protect from light) |
| SABC Dilution Buffer | 10ml | 4°C |
| TMB Substrate | 10ml | 4°C (Protect from light) |
| Stop Solution | 10ml | 4°C |
| Wash Buffer(25X) | 30ml | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipettetips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
| Uniprot | Q30154 |
| UniProt Protein Function: | Binds peptides derived from antigens that access the endocytic route of antigen presenting cells (APC) and presents them on the cell surface for recognition by the CD4 T-cells. The peptide binding cleft accommodates peptides of 10-30 residues. The peptides presented by MHC class II molecules are generated mostly by degradation of proteins that access the endocytic route, where they are processed by lysosomal proteases and other hydrolases. Exogenous antigens that have been endocytosed by the APC are thus readily available for presentation via MHC II molecules, and for this reason this antigen presentation pathway is usually referred to as exogenous. As membrane proteins on their way to degradation in lysosomes as part of their normal turn-over are also contained in the endosomal/lysosomal compartments, exogenous antigens must compete with those derived from endogenous components. Autophagy is also a source of endogenous peptides, autophagosomes constitutively fuse with MHC class II loading compartments. In addition to APCs, other cells of the gastrointestinal tract, such as epithelial cells, express MHC class II molecules and CD74 and act as APCs, which is an unusual trait of the GI tract. To produce a MHC class II molecule that presents an antigen, three MHC class II molecules (heterodimers of an alpha and a beta chain) associate with a CD74 trimer in the ER to form a heterononamer. Soon after the entry of this complex into the endosomal/lysosomal system where antigen processing occurs, CD74 undergoes a sequential degradation by various proteases, including CTSS and CTSL, leaving a small fragment termed CLIP (class-II-associated invariant chain peptide). The removal of CLIP is facilitated by HLA-DM via direct binding to the alpha-beta-CLIP complex so that CLIP is released. HLA-DM stabilizes MHC class II molecules until primary high affinity antigenic peptides are bound. The MHC II molecule bound to a peptide is then transported to the cell membrane surface. In B-cells, the interaction between HLA-DM and MHC class II molecules is regulated by HLA-DO. Primary dendritic cells (DCs) also to express HLA-DO. Lysosomal microenvironment has been implicated in the regulation of antigen loading into MHC II molecules, increased acidification produces increased proteolysis and efficient peptide loading. |
| NCBI Summary: | HLA-DRB5 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. DRB1 is expressed at a level five times higher than its paralogues DRB3, DRB4 and DRB5. The presence of DRB5 is linked with allelic variants of DRB1, otherwise it is omitted. There are 4 related pseudogenes: DRB2, DRB6, DRB7, DRB8 and DRB9. [provided by RefSeq, Jul 2008] |
| UniProt Code: | Q30154 |
| NCBI GenInfo Identifier: | 74754558 |
| NCBI Gene ID: | 3127 |
| NCBI Accession: | Q30154.1 |
| UniProt Secondary Accession: | Q30154,O00157, O00283, O46700, Q29703, Q29787, Q29788 Q30126, Q30150, Q30199, B2RBV6, C7C4X3, |
| UniProt Related Accession: | Q9TQE0,Q30154 |
| Molecular Weight: | 30kDa |
| NCBI Full Name: | HLA class II histocompatibility antigen, DR beta 5 chain |
| NCBI Synonym Full Names: | major histocompatibility complex, class II, DR beta 5 |
| NCBI Official Symbol: | HLA-DRB5 |
| NCBI Protein Information: | major histocompatibility complex, class II, DR beta 5 |
| UniProt Protein Name: | HLA class II histocompatibility antigen, DR beta 5 chain |
| UniProt Synonym Protein Names: | DR beta-5; DR2-beta-2; Dw2; MHC class II antigen DRB5 |
| Protein Family: | HLA class II histocompatibility antigen |
| UniProt Gene Name: | HLA-DRB5 |
*Note: Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Equilibrate the TMB substrate for at least 30 min at 37°C beforeuse. When diluting samples and reagents, they must be mixed completely andevenly. It is recommended to plot a standard curve for each test.
| Step | Protocol |
| 1. | Set standard, test sample and control (zero) wells on the pre-coatedplate respectively, and then, record their positions. It isrecommended to measure each standard and sample in duplicate. Washplate 2 times before adding standard, sample and control (zero) wells! |
| 2. | Add Sample and Biotin-detection antibody: Add 50µL of Standard, Blank or Sample per well. The blankwell is added with Sample Dilution Buffer. Immediately add 50 µL of biotin-labelled antibody workingsolution to each well. Cover with the plate sealer provided. Gently tap the plate to ensure thoroughmixing. Incubate for 45 minutes at 37°C. (Solutions are added to the bottom of micro-ELISA platewell, avoid touching plate walls and foaming). |
| 3. | Wash: Aspirate each well and wash, repeating the process three timesWash by filling each well with Wash Buffer (approximately 350µL)using a squirt bottle, multi-channel pipette, manifold dispenser orautomated washer. Complete removal of liquid at each step is essentialto good performance. After the last wash, remove any remaining WashBuffer by aspirating or decanting. Invert the plate and pat it againstthick clean absorbent paper. |
| 4. | HRP-Streptavidin Conjugate(SABC): Add 100µL of SABC workingsolution to each well. Cover with a new Plate sealer. Incubate for30minutes at 37°C. |
| 5. | Wash: Repeat the aspiration/wash process for five times. |
| 6. | TMB Substrate: Add 90µL of TMB Substrate to each well. Coverwith a new Plate sealer. Incubate for about 10-20 minutes at 37°C.Protect from light. The reaction time can be shortened or extendedaccording to the actual color change, but not more than 30minutes.When apparent gradient appeared in standard wells, you can terminatethe reaction. |
| 7. | Stop: Add 50µL of Stop Solution to each well. Color turn toyellow immediately. The adding order of stop solution should be as thesame as the substrate solution. |
| 8. | OD Measurement: Determine the optical density (OD Value) of each wellat once, using a microplate reader set to 450 nm. You should open themicroplate reader ahead, preheat the instrument, and set the testing parameters. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |