Anti-MSH2 Antibody (CAB1121)
- SKU:
- CAB1121
- Product type:
- Antibody
- Reactivity:
- Human
- Mouse
- Rat
- Host Species:
- Rabbit
- Isotype:
- IgG
- Antibody Type:
- Polyclonal Antibody
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Antibody Name: | Anti-MSH2 Antibody |
| Antibody SKU: | CAB1121 |
| Antibody Size: | 20uL, 50uL, 100uL |
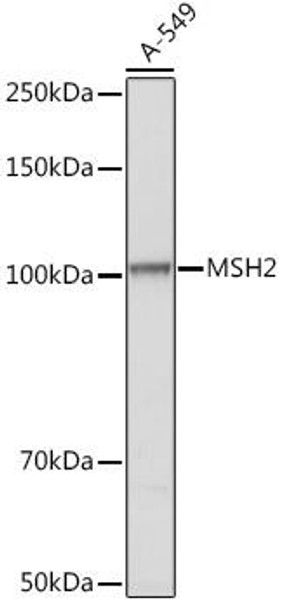
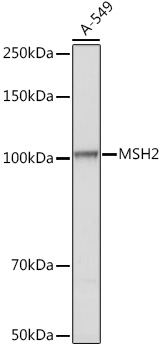
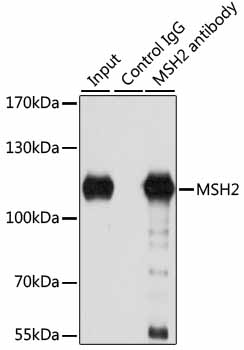
| Application: | WB IF IP |
| Reactivity: | Human, Mouse, Rat |
| Host Species: | Rabbit |
| Immunogen: | Recombinant fusion protein containing a sequence corresponding to amino acids 1-300 of human MSH2 (NP_000242.1). |
| Application: | WB IF IP |
| Recommended Dilution: | WB 1:500 - 1:2000 IF 1:50 - 1:200 IP 1:20 - 1:50 |
| Reactivity: | Human, Mouse, Rat |
| Positive Samples: | A-549 |
| Immunogen: | Recombinant fusion protein containing a sequence corresponding to amino acids 1-300 of human MSH2 (NP_000242.1). |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20°C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | MAVQ PKET LQLE SAAE VGFV RFFQ GMPE KPTT TVRL FDRG DFYT AHGE DALL AARE VFKT QGVI KYMG PAGA KNLQ SVVL SKMN FESF VKDL LLVR QYRV EVYK NRAG NKAS KEND WYLA YKAS PGNL SQFE DILF GNND MSAS IGVV GVKM SAVD GQRQ VGVG YVDS IQRK LGLC EFPD NDQF SNLE ALLI QIGP KECV LPGG ETAG DMGK LRQI IQRG GILI TERK KADF STKD IYQD LNRL LKGK KGEQ MNSA VLPE MENQ VAVS SLSA VIKF LELL SDDS NFGQ FELT TFDF SQYM |
| Gene ID: | 4436 |
| Uniprot: | P43246 |
| Cellular Location: | Nucleus |
| Calculated MW: | 97kDa/104kDa |
| Observed MW: | 25KDa |
| Synonyms: | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 |
| Background: | This locus is frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). When cloned, it was discovered to be a human homolog of the E. coli mismatch repair gene mutS, consistent with the characteristic alterations in microsatellite sequences (RER+ phenotype) found in HNPCC. Two transcript variants encoding different isoforms have been found for this gene. |
| UniProt Protein Function: | MSH2: Component of the post-replicative DNA mismatch repair system (MMR). Forms two different heterodimers: MutS alpha (MSH2- MSH6 heterodimer) and MutS beta (MSH2-MSH3 heterodimer) which binds to DNA mismatches thereby initiating DNA repair. When bound, heterodimers bend the DNA helix and shields approximately 20 base pairs. MutS alpha recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. MutS beta recognizes larger insertion-deletion loops up to 13 nucleotides long. After mismatch binding, MutS alpha or beta forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. In melanocytes may modulate both UV-B-induced cell cycle regulation and apoptosis. Heterodimer consisting of MSH2-MSH6 (MutS alpha) or MSH2- MSH3 (MutS beta). Both heterodimer form a ternary complex with MutL alpha (MLH1-PMS1). Interacts with EXO1. Part of the BRCA1- associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBS1 protein complex. This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains. Interacts with ATR. Interacts with SLX4/BTBD12; this interaction is direct and links MutS beta to SLX4, a subunit of different structure-specific endonucleases. Interacts with SMARCAD1. Ubiquitously expressed. Belongs to the DNA mismatch repair MutS family. |
| UniProt Protein Details: | Protein type:Tumor suppressor; DNA-binding Chromosomal Location of Human Ortholog: 2p21 Cellular Component: membrane; MutSalpha complex; MutSbeta complex; nuclear chromosome, telomeric region; nucleoplasm Molecular Function:ADP binding; ATP binding; ATPase activity; centromeric DNA binding; dinucleotide insertion or deletion binding; dinucleotide repeat insertion binding; DNA binding; double-strand/single-strand DNA junction binding; double-stranded DNA binding; enzyme binding; four-way junction DNA binding; guanine/thymine mispair binding; loop DNA binding; magnesium ion binding; mismatched DNA binding; MutLalpha complex binding; oxidized purine DNA binding; protein binding; protein C-terminus binding; protein homodimerization activity; protein kinase binding; single guanine insertion binding; single thymine insertion binding; single-stranded DNA binding; Y-form DNA binding Biological Process: B cell differentiation; B cell mediated immunity; cell cycle arrest; determination of adult life span; DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis; DNA repair; double-strand break repair; germ cell development; in utero embryonic development; intra-S DNA damage checkpoint; isotype switching; maintenance of DNA repeat elements; male gonad development; meiotic gene conversion; mismatch repair; negative regulation of DNA recombination; negative regulation of meiotic recombination; negative regulation of neuron apoptosis; oxidative phosphorylation; positive regulation of helicase activity; postreplication repair; response to UV-B; response to X-ray; somatic hypermutation of immunoglobulin genes; somatic recombination of immunoglobulin gene segments Disease: Lynch Syndrome I; Mismatch Repair Cancer Syndrome; Muir-torre Syndrome |
| NCBI Summary: | This locus is frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). When cloned, it was discovered to be a human homolog of the E. coli mismatch repair gene mutS, consistent with the characteristic alterations in microsatellite sequences (RER+ phenotype) found in HNPCC. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2012] |
| UniProt Code: | P43246 |
| NCBI GenInfo Identifier: | 1171032 |
| NCBI Gene ID: | 4436 |
| NCBI Accession: | P43246.1 |
| UniProt Secondary Accession: | P43246,O75488, B4E2Z2, |
| UniProt Related Accession: | P43246 |
| Molecular Weight: | 97,323 Da |
| NCBI Full Name: | DNA mismatch repair protein Msh2 |
| NCBI Synonym Full Names: | mutS homolog 2 |
| NCBI Official Symbol: | MSH2 |
| NCBI Official Synonym Symbols: | FCC1; COCA1; HNPCC; LCFS2; HNPCC1 |
| NCBI Protein Information: | DNA mismatch repair protein Msh2 |
| UniProt Protein Name: | DNA mismatch repair protein Msh2 |
| UniProt Synonym Protein Names: | MutS protein homolog 2 |
| Protein Family: | DNA mismatch repair protein |
| UniProt Gene Name: | MSH2 |
| UniProt Entry Name: | MSH2_HUMAN |