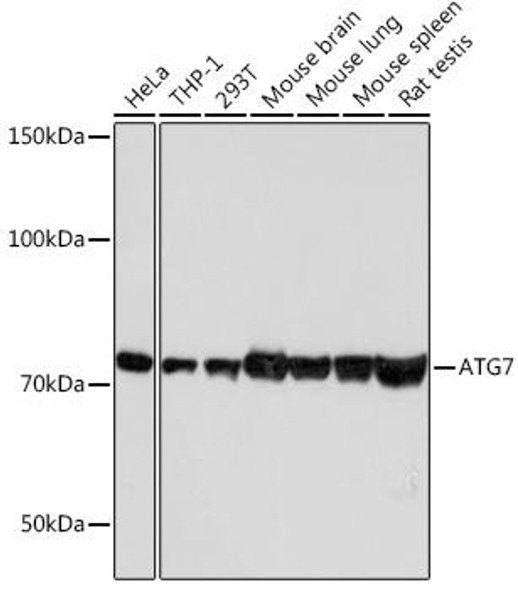
| Background: | This gene encodes an E1-like activating enzyme that is essential for autophagy and cytoplasmic to vacuole transport. The encoded protein is also thought to modulate p53-dependent cell cycle pathways during prolonged metabolic stress. It has been associated with multiple functions, including axon membrane trafficking, axonal homeostasis, mitophagy, adipose differentiation, and hematopoietic stem cell maintenance. Alternative splicing results in multiple transcript variants. |
| UniProt Protein Function: | ATG7: Functions as an E1 enzyme essential for multisubstrates such as ATG8-like proteins and ATG12. Forms intermediate conjugates with ATG8-like proteins (GABARAP, GABARAPL1, GABARAPL2 or MAP1LC3A). PE-conjugation to ATG8-like proteins is essential for autophagy. Also acts as an E1 enzyme for ATG12 conjugation to ATG5 and ATG3. Homodimer. Interacts with ATG3 and ATG12. The complex, composed of ATG3 and ATG7, plays a role in the conjugation of ATG12 to ATG5. Widely expressed, especially in kidney, liver, lymph nodes and bone marrow. Belongs to the ATG7 family. 2 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:Ubiquitin conjugating system; Autophagy Chromosomal Location of Human Ortholog: 3p25.3 Cellular Component: cytoplasm; axoneme; cytosol Molecular Function:protein binding; protein homodimerization activity; APG8 activating enzyme activity; APG12 activating enzyme activity; ubiquitin activating enzyme activity; transcription factor binding Biological Process: regulation of protein ubiquitination; positive regulation of apoptosis; protein ubiquitination; pyramidal neuron development; adult walking behavior; post-embryonic development; protein transport; cardiac muscle cell development; protein amino acid lipidation; protein catabolic process; positive regulation of autophagy; amino acid metabolic process; central nervous system neuron axonogenesis; cerebellar Purkinje cell layer development; protein modification process; C-terminal protein lipidation; liver development; cellular response to starvation; neurological system process; cellular response to nitrogen starvation; protein modification by small protein conjugation; positive regulation of protein modification process; positive regulation of protein catabolic process; mitochondrion degradation; cerebral cortex development; positive regulation of macroautophagy; negative regulation of apoptosis |
| NCBI Summary: | This gene was identified based on homology to Pichia pastoris GSA7 and Saccharomyces cerevisiae APG7. In the yeast, the protein appears to be required for fusion of peroxisomal and vacuolar membranes. The protein shows homology to the ATP-binding and catalytic sites of the E1 ubiquitin activating enzymes. [provided by RefSeq, Jan 2009] |
| UniProt Code: | O95352 |
| NCBI GenInfo Identifier: | 62286592 |
| NCBI Gene ID: | 10533 |
| NCBI Accession: | O95352.1 |
| UniProt Secondary Accession: | O95352,Q7L8L0, Q9BWP2, Q9UFH4, B4E170, E9PB95, |
| UniProt Related Accession: | O95352 |
| Molecular Weight: | 68,618 Da |
| NCBI Full Name: | Ubiquitin-like modifier-activating enzyme ATG7 |
| NCBI Synonym Full Names: | autophagy related 7 |
| NCBI Official Symbol: | ATG7 |
| NCBI Official Synonym Symbols: | GSA7; APG7L; APG7-LIKE |
| NCBI Protein Information: | ubiquitin-like modifier-activating enzyme ATG7; hAGP7; autophagy-related protein 7; ATG12-activating enzyme E1 ATG7; ATG7 autophagy related 7 homolog; ubiquitin activating enzyme E1-like protein; ubiquitin-activating enzyme E1-like protein |
| UniProt Protein Name: | Ubiquitin-like modifier-activating enzyme ATG7 |
| UniProt Synonym Protein Names: | ATG12-activating enzyme E1 ATG7; Autophagy-related protein 7; APG7-like; hAGP7; Ubiquitin-activating enzyme E1-like protein |
| Protein Family: | Ubiquitin-like modifier-activating enzyme |
| UniProt Gene Name: | ATG7 |
| UniProt Entry Name: | ATG7_HUMAN |