Anti-APEX2 Antibody (CAB16116)
- SKU:
- CAB16116
- Product type:
- Antibody
- Reactivity:
- Human
- Host Species:
- Rabbit
- Isotype:
- IgG
- Antibody Type:
- Polyclonal Antibody
- Research Area:
- Cell Cycle
Frequently bought together:
Description
| Antibody Name: | Anti-APEX2 Antibody |
| Antibody SKU: | CAB16116 |
| Antibody Size: | 20uL, 50uL, 100uL |
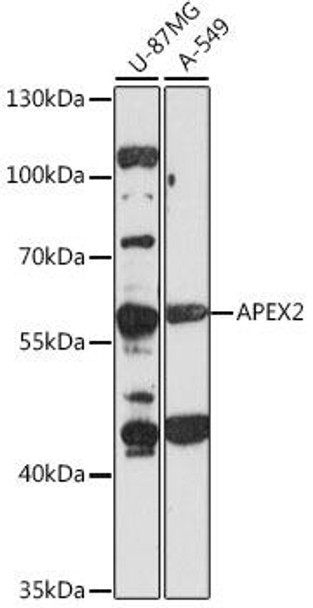
| Application: | WB |
| Reactivity: | Human |
| Host Species: | Rabbit |
| Immunogen: | Recombinant fusion protein containing a sequence corresponding to amino acids 1-180 of human APEX2 (NP_055296.2). |
| Application: | WB |
| Recommended Dilution: | WB 1:500 - 1:2000 |
| Reactivity: | Human |
| Positive Samples: | U-87MG, A-549 |
| Immunogen: | Recombinant fusion protein containing a sequence corresponding to amino acids 1-180 of human APEX2 (NP_055296.2). |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20°C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | MLRV VSWN INGI RRPL QGVA NQEP SNCA AVAV GRIL DELD ADIV CLQE TKVT RDAL TEPL AIVE GYNS YFSF SRNR SGYS GVAT FCKD NATP VAAE EGLS GLFA TQNG DVGC YGNM DEFT QEEL RALD SEGR ALLT QHKI RTWE GKEK TLTL INVY CPHA DPGR PERL VFKM RFYR LLQI |
| Gene ID: | 27301 |
| Uniprot: | Q9UBZ4 |
| Cellular Location: | Cytoplasm, Mitochondrion, Nucleus |
| Calculated MW: | 57kDa |
| Observed MW: | 57kDa |
| Synonyms: | APEX2, APE2, APEXL2, XTH2, ZGRF2 |
| Background: | Apurinic/apyrimidinic (AP) sites occur frequently in DNA molecules by spontaneous hydrolysis, by DNA damaging agents or by DNA glycosylases that remove specific abnormal bases. AP sites are pre-mutagenic lesions that can prevent normal DNA replication so the cell contains systems to identify and repair such sites. Class II AP endonucleases cleave the phosphodiester backbone 5' to the AP site. This gene encodes a protein shown to have a weak class II AP endonuclease activity. Most of the encoded protein is located in the nucleus but some is also present in mitochondria. This protein may play an important role in both nuclear and mitochondrial base excision repair. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. |
| UniProt Protein Function: | APEX2: Function as a weak apurinic/apyrimidinic (AP) endodeoxyribonuclease in the DNA base excision repair (BER) pathway of DNA lesions induced by oxidative and alkylating agents. Initiates repair of AP sites in DNA by catalyzing hydrolytic incision of the phosphodiester backbone immediately adjacent to the damage, generating a single-strand break with 5'-deoxyribose phosphate and 3'-hydroxyl ends. Displays also double-stranded DNA 3'-5' exonuclease, 3'-phosphodiesterase activities. Shows robust 3'-5' exonuclease activity on 3'-recessed heteroduplex DNA and is able to remove mismatched nucleotides preferentially. Shows fairly strong 3'-phosphodiesterase activity involved in the removal of 3'-damaged termini formed in DNA by oxidative agents. In the nucleus functions in the PCNA-dependent BER pathway. Required for somatic hypermutation (SHM) and DNA cleavage step of class switch recombination (CSR) of immunoglobulin genes. Required for proper cell cycle progression during proliferation of peripheral lymphocytes. Belongs to the DNA repair enzymes AP/ExoA family. |
| UniProt Protein Details: | Protein type:Lyase; EC 4.2.99.18; Mitochondrial Chromosomal Location of Human Ortholog: Xp11.21 Cellular Component: intracellular membrane-bound organelle; mitochondrial inner membrane; nucleolus; nucleus Molecular Function:DNA-(apurinic or apyrimidinic site) lyase activity; DNA binding; zinc ion binding; double-stranded DNA specific 3'-5' exodeoxyribonuclease activity Biological Process: base-excision repair; DNA catabolic process, exonucleolytic; cell cycle; DNA catabolic process, endonucleolytic; DNA recombination |
| NCBI Summary: | Apurinic/apyrimidinic (AP) sites occur frequently in DNA molecules by spontaneous hydrolysis, by DNA damaging agents or by DNA glycosylases that remove specific abnormal bases. AP sites are pre-mutagenic lesions that can prevent normal DNA replication so the cell contains systems to identify and repair such sites. Class II AP endonucleases cleave the phosphodiester backbone 5' to the AP site. This gene encodes a protein shown to have a weak class II AP endonuclease activity. Most of the encoded protein is located in the nucleus but some is also present in mitochondria. This protein may play an important role in both nuclear and mitochondrial base excision repair. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Nov 2012] |
| UniProt Code: | Q9UBZ4 |
| NCBI GenInfo Identifier: | 73921676 |
| NCBI Gene ID: | 27301 |
| NCBI Accession: | Q9UBZ4.1 |
| UniProt Secondary Accession: | Q9UBZ4,Q9Y5X7, |
| UniProt Related Accession: | Q9UBZ4 |
| Molecular Weight: | 57,401 Da |
| NCBI Full Name: | DNA-(apurinic or apyrimidinic site) lyase 2 |
| NCBI Synonym Full Names: | APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| NCBI Official Symbol: | APEX2 |
| NCBI Official Synonym Symbols: | APE2; XTH2; ZGRF2; APEXL2 |
| NCBI Protein Information: | DNA-(apurinic or apyrimidinic site) lyase 2; DNA-(apurinic or apyrimidinic site) lyase 2; AP endonuclease 2; AP endonuclease XTH2; zinc finger, GRF-type containing 2; apurinic/apyrimidinic endonuclease-like 2 |
| UniProt Protein Name: | DNA-(apurinic or apyrimidinic site) lyase 2 |
| UniProt Synonym Protein Names: | AP endonuclease XTH2; APEX nuclease 2; APEX nuclease-like 2; Apurinic-apyrimidinic endonuclease 2; AP endonuclease 2 |
| Protein Family: | Aminopeptidase |
| UniProt Gene Name: | APEX2 |
| UniProt Entry Name: | APEX2_HUMAN |